Tutorial
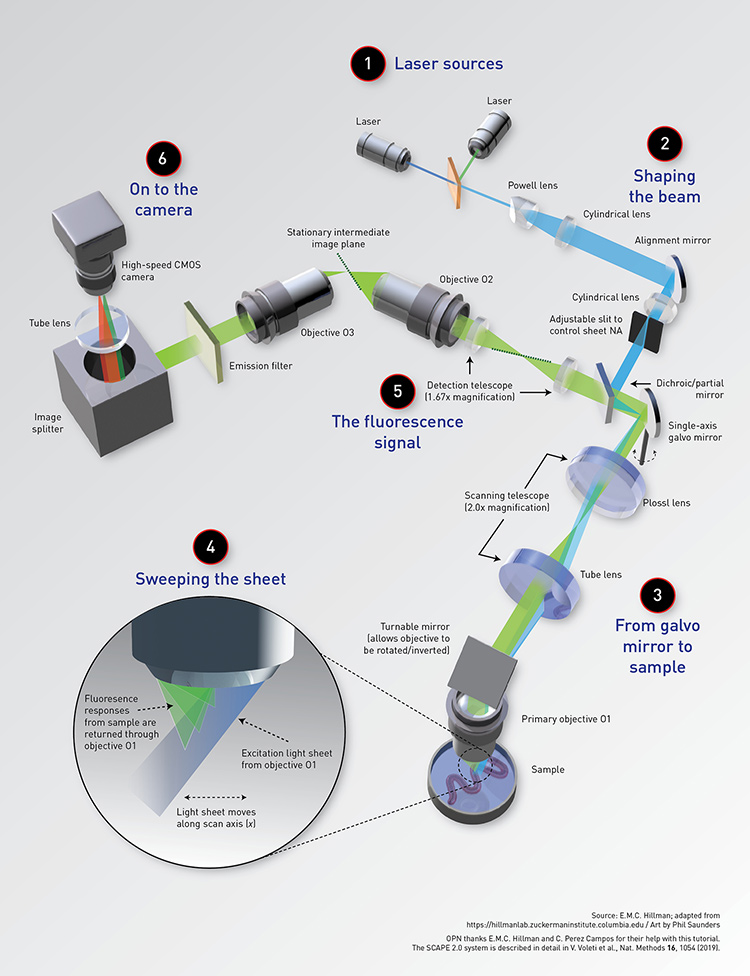
SCAPE 2.0 Microscopy
Speeding up light-sheet imaging of live biological specimens.
[Enlarge graphic] Source: E.M.C. Hillman; adapted from https://hillmanlab.zuckermaninstitute.columbia.edu / Art by Phil Saunders
…Log in or become a member to view the full text of this article.
This article may be available for purchase via the search at Optica Publishing Group.
Optica Members get the full text of Optics & Photonics News, plus a variety of other member benefits.