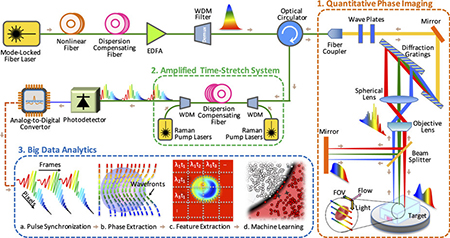
TS-QPI and analytics system. [Credit: Sci. Rep., doi: 10.1038/srep21471] [Enlarge image]
OSA Fellow Bahram Jalali and his colleagues at the University of California, Los Angeles (UCLA; USA) have proposed a new time-stretch quantitative phase imaging (TS-QPI) system for capturing individual biological cells, integrated with deep machine-learning for cell classification (Sci. Rep., doi: 10.1038/srep21471). Corresponding author Claire Lifan Chen said that the technique “achieves record high-accuracy in label-free cell classification” without the potentially damaging chemical stains typically used in cell analysis. The technique could, the authors say, be used in a wide variety of applications, including data-driven cancer diagnostics, drug development, personalized genomics and biofuel production.
Time-stretch image capture
TS-QPI builds on the team’s previous work on amplified time-stretch dispersive Fourier transform (Nat. Photon., doi: 10.1038/nphoton.2012.359). With TS-QPI, an individual cell’s spatial information is encoded in the spectrum of laser pulses within a pulse-duration of sub-nanoseconds. Each pulse is then stretched in time and digitized in real time by an electronic analog-to-digital converter. The UCLA scientists report being able to capture blur-free phase and intensity images of cells at a rate of 100,000 cells per second with this new technique. (For comparison, existing flow-cytometry camera setups capture 2,000 cells per second.)
Crunching big data
TS-QPI produces massive, information-rich image datasets. The team leveraged these datasets to create an algorithm for deep machine-learning to extract 16 different features from images of individual cells—e.g., refractive index, absorption and diameter—and classify the cells in a “hyperdimensional space” composed of these 16 features. The authors say that this is the first time deep machine-learning, commonly used in speech processing and image recognition, has been used in label-free classification of cells.
Jalali and his team conducted two experimental demonstrations of TS-QPI: classifying white blood cells against colon cancer cells, and classifying lipid-accumulating algal strains for biofuel production. Their results show that compared to classification by individual biophysical parameters, their label-free TS-QPI technique improved detection accuracy from 77.8 percent to 95.5 percent.
The UCLA team worked in collaboration with scientists from NantWorks, USA, who sponsored the study.
