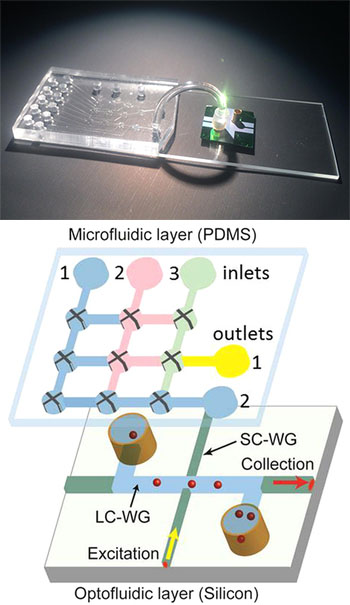
(Top) The research team’s hybrid device integrates a microfluidic chip for sample preparation (left) and an optofluidic chip for detection of individual Ebola RNA molecules (right). A connecting tube allows the prepared sample to pass into the optical detection system. [Image: Joshua Parks] (Bottom) Schematic diagram of the microfluidic-optofluidic setup. [Image: Cai et al., Sci. Reports, doi: 10.1038/srep14494]
Scientists at four U.S. institutions have developed a chip-based device that uses optical detection to sense infection by the deadly Ebola virus (Sci. Reports, doi: 10.1038/srep14494). The device, which ingeniously combines microfluidics and optics in a modular setup, reportedly can achieve sensitivities and dynamic range comparable to those of commonly used, far more complex and time-consuming lab techniques.
Those advantages, in the view of the research team, could make the technology a strong contender as the basis for portable instruments, to provide rapid, early detection of the virus in field settings.
The search for rapid detection
The outbreak of Ebola, a deadly hemorrhagic fever, in West Africa has claimed more than 11,000 lives in the past two years. It has also spotlighted the need for rapid, accurate diagnosis, since the disease tends to present with maddeningly nonspecific symptoms—such as fever and headache—in its early stages, before the virus has gained a strong foothold in the body. And, as many cases have arisen in resource-poor countries, detectors for Ebola ideally need to be cheap, compact and ready to be deployed in the field rather than in a lab.
At present, the most reliable test for Ebola involves the technique of reverse-transcriptase polymerase chain reaction (RT-PCR), in which genetic material from the virus is amplified until there’s enough to provide a detectable signal. RT-PCR can sniff out Ebola with high sensitivity and specificity. But the amplification step makes the process complex and unwieldy, requiring a lab setting and making it a poor candidate for point-of-care use.
A modular chip-based approach
A team led by OSA Fellow Holger Schmidt of the University of California, Santa Cruz—with colleagues there and at Brigham Young University, Utah, Texas Biomedical Institute, and the University of California, Berkeley—took up the challenge to create a more field-ready, chip-scale device. The team’s system combines microfluidics and optical waveguide technology. And, to circumvent the compatibility problems that sometimes arise in combining such systems, the scientists built the microfluidic and optical systems on different, interconnected chips.
The first, microfluidic layer handles isolation and concentration of Ebola virus RNA. The sample to be tested is first exposed to a solution containing magnetic microbeads, to which a short nucleotide fragment, selected for its affinity for Ebola RNA, has been attached. The nucleotide strip grabs onto the nearest Ebola RNA it can find suspended in the sample, and attaches the suspect RNA to the magnetic bead.
The solution is then pumped into a series of tiny polydimethylsiloxane channels on the microfluidic chip, where a magnet pulls down and holds the Ebola-bearing microbeads in place, effectively isolating the Ebola while the rest of the solution washes through. The Ebola RNA fragments are then thermally released from the microbeads and labeled with a fluorescent molecule.
The tagged RNA fragments next flow through a connecting tube into the optofluidic layer. That layer consists of a chip with two perpendicular waveguides: a microchannel liquid-core guide that serves double duty as a carrier of sample material and optical signal; and a solid-core waveguide tied to an excitation laser. As the tagged RNA snippets pass through the intersection point of the two waveguides, they are zapped by the excitation laser, and the resulting fluorescent signal passes through the liquid-core waveguide and is detected and counted at the channel’s end.
High specificity and dynamic range
The system avoids the time-consuming, complex amplification step of RT-PCR, by reading the signal directly from individual tagged RNAs. Yet the team reports that, in its experiments, the setup rivaled RT-PCR in sensitivity, and was capable of picking up an accurate signal across six orders of magnitude—or more, when an on-chip pre-concentration step was added. That, says team leader Schmidt, covers “the entire range of what would be seen in an infected person.” What’s more, the chip-based system was completely specific to Ebola; tests with other closely related hemorrhagic-fever viruses used as controls came up negative.
The next step, according to Schmidt, will be end-to-end testing with raw blood samples, which must take place at a biosafety level 4 facility. The team is also looking at extending the system to detecting other pathogens.
