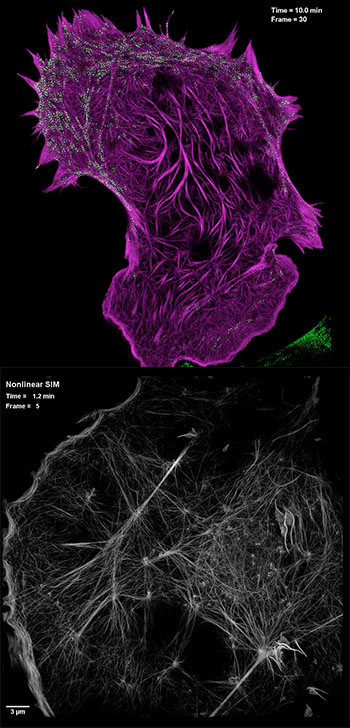
(Top) Still image from video showing interaction of cytoskeletal actin filliaments (purple) with the cellular motor protein myosin (green), captured by high-NA SIM. (Bottom) Still image from video showing actin cytoskeletal detail using PA NL-SIM. [Images: Betzig Lab, HHMI/Janelia Research Campus]
Scientists from the United States and China, led by 2014 Nobel laureate Eric Betzig of the Howard Hughes Medical Institute (USA), have squeezed 50 percent better resolution from a key microscopy technique for imaging processes in living cells (Science, doi: 10.1126/science.aab3500). The new techniques, according to the team, fill “an unmet need for minimally invasive tools” in a key area of biological microscopy—and have already been used to answer some long-standing questions in protein dynamics.
Super-resolution’s Achilles heel
Super-resolution (SR) fluorescence microscopy, the development of which snagged the 2014 Nobel Prize in Chemistry, uses laser and fluorescence techniques to blast through the roughly 200-nm diffraction-limited spatial resolution of conventional light microscopes, by as much as an order of magnitude. But while it’s been a revolutionary tool for studying biological macromolecules, SR microscopy has several pitfalls in looking at actual protein dynamics, where many of biology’s most interesting problems lie.
Perhaps the biggest issue is phototoxicity: the high levels of illumination and strong concentrations of fluorescent markers required by most SR techniques can damage live cells after even short periods, wiping out the very dynamics being studied. And some SR microscopy techniques embody processes of switching on and off fluorescent molecules that operate too slowly to illuminate protein processes, which can happen at blazing speeds.
Toward a sharper, faster SIM
One SR technique that avoids these pitfalls is structured-illumination microscopy (SIM), under which the sample is illuminated by multiple different light patterns (analogous to bar codes) and the resulting images are reconstructed computationally into detailed 3-D images. SIM requires orders of magnitude less light than other SR techniques, limiting phototoxicity, and can also proceed much faster, with the potential of capturing dynamic protein processes. But unfortunately, conventional SIM can get only to around 100-nm resolution—twice the diffraction limit of conventional light microscopes, but well below the resolution offered by the most powerful SR techniques.
Betzig’s team attacked the resolution limits of SIM using two approaches. First, the scientists used an ultrahigh numerical aperture (NA) lens on a conventional light microscope, combined with a variant of SIM that relies on total internal reflection fluorescence (high-NA TIRF SIM), to push the resolution limit down to 84 nm, with subsecond acquisition speeds, for both high spatial and temporal resolution.
In a second approach, the researchers exploited a newly developed, reversibly photoswitchable fluorescent protein in a technique called patterned-activation nonlinear SIM (PA NL-SIM) to push down spatial resolution even further. Nonlinear SIM relies on high-harmonic generation from the patterned light that can be used to extend the resolution. The team was able to improve on that technique using the newly developed protein, and a strategic, selective approach to turning on and off only the subset of fluorescent molecules in the cell required for a clear image. The result was dramatically reduced stress on the cell, and spatial resolutions of 45 to 62 nm—again at the subsecond timescales needed to capture protein processes.
Catching proteins in the act
Leveraging these two improvement’s, Betzig’s team has shed new light on some biological questions previously outside of the realm of direct observation. The scientists were able to image the growth of clathrin-coated pits, a key structure in the process known as endocytosis, in which cells engulf other molecules; the dynamics of the cytoskeleton; and the transport of internal cellular cargoes to and from the Golgi apparatus. And combining the high-NA approach, which allows the tracking of multiple, different colored fluorescent proteins, with the higher resolution offered by PA NL-SIM let the team image the movement of multiple proteins at a spatial resolution greater than the high-NA approach alone would have allowed.
The combination of better spatial resolution and higher temporal resolution, the study concludes, fills “an important gap between the 100-nm limit of traditional SIM and the macromolecular level of localization microscopy”—potentially in the sweet spot for capturing detailed protein dynamics. Says Betzig: “This will bring super-resolution to live-cell imaging for real.”
