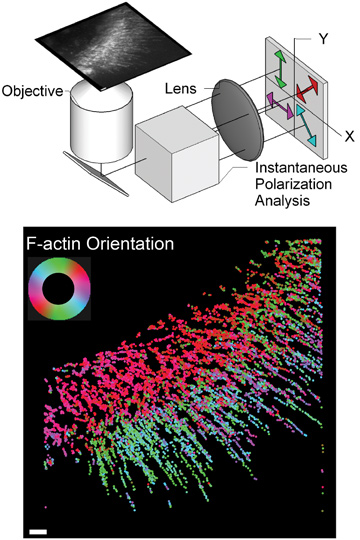
 (Top) Simplified schematic of instantaneous fluorescence polscope. (Bottom) Molecular orientation of F-actin network at the leading edge of migrating cell. Color-coded according to the color-wheel in the inset; scale bar = 1 µm.
(Top) Simplified schematic of instantaneous fluorescence polscope. (Bottom) Molecular orientation of F-actin network at the leading edge of migrating cell. Color-coded according to the color-wheel in the inset; scale bar = 1 µm.
Molecular order underpins many directed functions performed by cells. For example, ordered assemblies of actin cytoskeleton and myosin motor proteins generate contractile forces necessary for directed cell migration and cell division. Despite molecular order’s importance across biology and soft-matter physics, its measurement in live cells and dynamic systems, across spatial and temporal scales, remains challenging. Molecular order is typically too fine to visualize with super-resolution light microscopes. Electron microscopes can resolve molecular order, but only offer a snapshot of the ordered organization in fixed specimens.
We have developed an optical microscope and computational methods that reveal the molecular orientation of single molecules and their ensembles in live cells—with a temporal resolution of 20 Hz, localization accuracy of 50 nm and orientation accuracy of less than 5 degrees.1 The microscope has enabled the study of some outstanding biological questions that hinge on how nanoscale molecules come together to form dynamic micron-scale assemblies in cells.
Our microscope, called an instantaneous fluorescence polscope, exploits the photophysics of fluorophores, which emit polarized radiation in a manner akin to nanoscale dipole antennas. Our microscope excites all fluorophores in a sample equally, irrespective of their 3-D orientation, and detects the polarization of emission along four orientations in a single snapshot.1 Analysis of polarized emission allows determination of the fluorophore’s orientation and, in turn, the orientation of the molecule containing it.
We also developed algorithms for automatically detecting and tracking the position, orientation and intensity of single fluorescent particles in live cells, using movies acquired with the instantaneous fluorescence polscope. This computational-imaging approach allowed us to visualize flow and molecular orientation of filamentous actin (F-actin) at the leading edge of migrating cells. Before this work, imaging the orientation of F-actin required electron microscopy and was therefore limited to fixed (nonliving) cells. Our approach reveals the flow speed and molecular orientation of F-actin simultaneously, and thus allows for correlative analysis in live cells.
Our microscope has led to new insights into biological questions. In collaboration with the Whitman center at the Marine Biological Laboratory, Woods Hole, Mass., USA, we measured the assembly kinetics of septin intermediate filaments, which play an important role in cell shape, cell migration and cell division by forming assemblies of diverse shapes.1 We found that septin subunits in live cells are more dynamic than previously thought. In a more recent, multi-institutional collaboration, we have used the instantaneous polscope to explore the dynamics of the flow within cells of integrin proteins, which transmit the force generated by actin.
Researchers
Shalin B. Mehta, Marine Biological Laboratory, Woods Hole, Mass., USA, and University of Chicago, Chicago, Ill., USA
Rudolf Oldenbourg, Marine Biological Laboratory and Brown University, Providence, R.I., USA
Tomomi Tani, Marine Biological Laboratory
Reference
1. S.B. Mehta et al. Proc. Natl. Acad. Sci. USA, doi: 10.1073/pnas.1607674113 (2016).
