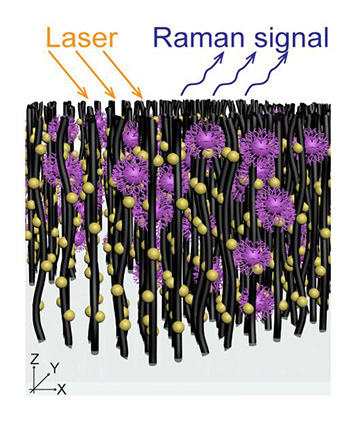
In the Penn State group’s VIRRION system, an array of nanotubes decorated with gold nanoparticles captures virus molecules, the strains of which are then identified using Raman spectroscopy. [Credit: Terrones Lab/Penn State]
Discovering and identifying viruses plays a large part in controlling or even preventing disease outbreaks. A team led by researchers at Penn State University, USA, has developed a handheld microfluidic platform that can optically identify multiple virus strains in only a few minutes (PNAS, doi: 10.1073/pnas.1910113117). The device, according to the team, shows promise as a rapid, portable alternative to current methods of virus surveillance, which require laboratory infrastructure and take hours or days to produce results.
Keeping tabs on the enemy
Globalization has drastically altered the spread and transmission of infectious diseases. Globe-spanning travel and trade mean that viruses and other infectious agents are no longer restricted to one host species or region. Effective surveillance systems, particularly in regions of the developing world, remain crucial for identifying disease patterns and ultimately containing dangerous viral outbreaks like Zika and Ebola.
“Viruses evolve rapidly and unpredictably, challenging the effectiveness of disease diagnostics,” said Yin-Ting (Tim) Yeh, the new study’s corresponding co-author. “To help control outbreaks and understand their origins, the first step is often isolating viruses from infected samples for characterization.”
A CNT virus filter
Instead of time-consuming, lab-based methods, Yeh and his colleagues focused on creating a faster way to identify viruses using carbon nanotubes (CNTs) and optical techniques. Their device, called VIRRION, consists of aligned nitrogen-doped CNT arrays in a herringbone pattern. Separate zones of the herringbone arrays are engineered to have intertubular distances ranging from 22 to 720 nm, to match the sizes of different viruses. VIRRION thus acts as a filtration device, capturing individual viruses by size while preserving their structural integrity.
Next, the researchers functionalized the CNTs with gold nanoparticles to enable surface-enhanced Raman spectroscopy to optically identify the virus particles. They validated the platform using different subtypes of avian influenza virus, as well as human respiratory viruses. For the latter, VIRRION successfully captured and identified viruses in nasal swab samples from patients who had been diagnosed with rhinovirus, influenza A virus or human parainfluenza virus type 3.
Machine learning
“We implemented a machine-learning protocol to match the spectra to different types and subtypes of viruses, showing structural differences in the envelope proteins and RNA material,” said Mauricio Terrones, the study’s other corresponding co-author. “This combination allowed us to identify viruses with 90% accuracy using Raman spectroscopy within minutes.”
As a next step, Terrones and Yeh wish to pursue commercialization of the technology as an effective disease-monitoring system and real-time virus identification tool. However, VIRRION still needs some additional upgrades before being ready for the field. The team is now working on improving the identification accuracy and the Raman signal using novel materials and approaches.
In addition to researchers at Penn State University, the team also included scientists from New York University, USA.
