If you could simply look at a bacterial colony and identify it, what would that mean to the worlds of medicine, food safety and biodefense? For years, microbiologists have used informed observations of bacterial colonies—noting their shape, size, color, texture and odor—in order to gain insight into the organism’s identity. However, these assessments do not provide enough information for a quantitative and conclusive identification. Therefore, lengthy biochemical and genetic evaluations are the next steps in virtually all laboratories.
New elastic light scatter (ELS) techniques may simplify that process. They can provide a low-cost way to quickly identify harmful bacteria by analyzing the unique scattering patterns that these organisms produce when scanned by a laser. By linking ELS pattern databases, scientists could compare unknown patterns with laboratories all over the world and hopefully find a match within seconds.
The desire to create rapid bacteria identification systems is by no means new. For over a century, scientists have worked to protect people from disease-causing microorganisms. Thanks to these efforts, there are many food preparation and delivery methods in place to ensure that our food remains safe until it reaches our refrigerators and pantries. Water supply systems have also been designed to remove contaminating organisms to a point well below their infective doses.
 Photon-colony interaction changes wavefront characteristic. The interaction between photons and a bacterial colony alters wavefront properties based on the bacterial colony’s physical characteristics. (Top) 3-D diagram showing the principle of the bacterial scatterometer (i.e., ELS). (Bottom) The concept of forward scatterometry from a bacterial colony.
Photon-colony interaction changes wavefront characteristic. The interaction between photons and a bacterial colony alters wavefront properties based on the bacterial colony’s physical characteristics. (Top) 3-D diagram showing the principle of the bacterial scatterometer (i.e., ELS). (Bottom) The concept of forward scatterometry from a bacterial colony.
More recently, scientists have been looking for much more rapid pathogen detection options to help manage the potential impact of bioterrorist acts that could endanger military forces, critical service industries or the community at large. In all cases, the earlier the organism is identified, the better.
What is “early” bacteria identification?
What is considered “early” in the identification of bacteria? The answer depends on where you are and what you are looking for. In addition, various techniques vastly differ in their speed, accuracy and practical implementation.
The food industry
In the food industry, foodborne pathogens would ideally be caught prior to the processing or shipment of large volumes of food. This might be within 24 hours of harvest or any time before the food has reached consumers. Early identification significantly reduces the impact of costly recalls and the chance of public health problems.
Clinical settings
In the medical world, rapid identification of pathogens results in faster and more accurate patient treatment. In addition, if the identity of the bacteria can be quickly matched with antibiotic sensitivity, there will be less of a possibility that clinicians will prescribe inappropriate antibiotics—which contributes to a troubling increase in drug-resistant bacteria. A target for early identification in this scenario would be within 12 to 24 hours after a pathogen has been found to be present.
Biodefense applications
In the area of biodefense, detection time again depends upon the nature of the threat. For example, the presence of a normal or genetically modified pathogen is unlikely to be noticed until clear evidence of a problem emerges, such as a sudden unexplained increase in hospitalizations. This could take 24 to 48 hours or more after exposure.
Current methods of identifying pathogens
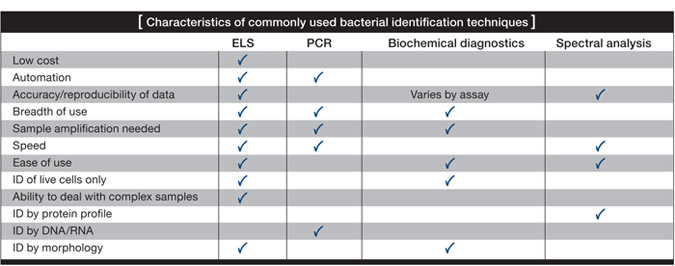
Currently, three fundamental approaches are used to detect pathogens: 1) biochemical identification, which is typically used in the food and clinical industries; 2) genetic tools such as polymerase chain reaction (PCR) and DNA fingerprinting, which are also used in food and clinical industries, as well as for biodefense; and lastly, 3) spectral analysis techniques such as mass spectrometry, which has recently become a presence in clinical laboratories.
With perhaps the exception of spectral analysis, these approaches take a considerable amount of time. Even PCR, which is considered the fastest technique and the one favored by industries and militaries, almost always requires some growth of the sample in order to ensure accurate identification.
The amplification time depends on the number and growth rate of microorganisms in the sample. It may be four to 12 hours before bacteria numbers reach a level useful for these identification tools. Ultimately, if you cannot increase the number of organisms, you will not identify your sample.
|
|
|---|
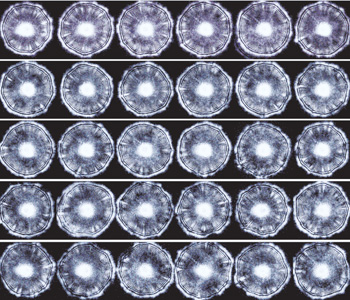 Forward-scatter patterns generated using ELS. ELS patterns from different colonies of Listeria innocua on a single 96-well plate. These forward-scatter patterns or phenotypes are highly reproducible. Scans were made using a fully automated robotic system.
Forward-scatter patterns generated using ELS. ELS patterns from different colonies of Listeria innocua on a single 96-well plate. These forward-scatter patterns or phenotypes are highly reproducible. Scans were made using a fully automated robotic system.
The optics behind ELS
ELS works by encoding the micro- and macro-structural morphology of the colony onto an interrogating wavefront. The colony’s optical secrets are then decoded by examining the forward-scattering pattern resulting from the wavefront propagation towards the far field. The use of a spatial light modulator (SLM) and a liquid-crystal display represents a renowned technique in optical engineering to control wavefront modulation. We can understand the photon-colony interaction as a “biological SLM” that changes the wavefront characteristics based on the colony’s physical differences.
With high coherency and nanometer wavelength, the incoming plane wave passes through the bacterial colony from top to bottom. Various physical parameters—such as refractive indices, the local density of bacteria and the individual shape of bacteria—can all influence the incoming photons. This interaction through the z-depth of the colony accumulates and finally disrupts the amplitude and phase of the incoming photons.
Before the photons interact with the colonies, the spatial distribution of the incoming wavefront is a plane wave with typical circular Gaussian intensity distribution. However, upon leaving the far side of the colonies, the departing wavefront is now encoded with different amplitude and phase modulation in 2-D space. If we apply the Huygens-Fresnel principle in rectangular coordinates, the process (1)-(3) of the image above can be modeled as:
|
|
|---|
where xi,yi are points in the image plane, Σ denotes the colony surface, t(xa,ya) is the 2-D transmission coefficient, E1(xa,ya) is the 2-D incident Gaussian beam, φ(xa,ya) is the 2-D phase-modulation factor, rai is the distance from aperture plane to image plane, λ is the wavelength and θ is the angle of rai to the optical axis. When this disturbed wavefront is propagated to the imaging plane, the characteristic scattering pattern is generated via spatial interference.
ELS equipment
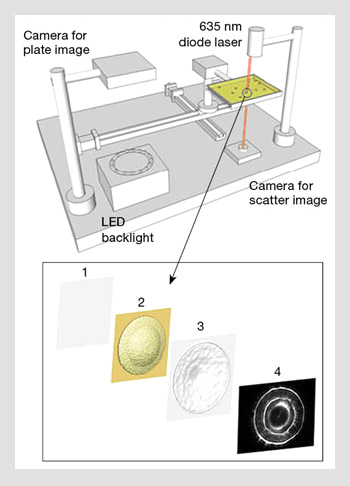
The optical setup for ELS consists of two major sections: a plate reader and a forward scatterometer. The plate reader determines the spatial distribution of individual bacterial colonies by measuring the transmitted light from the LED backlight. Both the location and the individual characteristics of a bacterial colony (diameter, circularity, etc.) can be extracted by image-processing algorithms. The plate is then transferred to the forward scatterometer for laser interrogation. The incoming 635-nm laser beam with a 1/e2 of 1 mm is directed through each single colony, and the associated scattering pattern is captured.
Two major points must be addressed in this process. First, since the typical number of colonies on a single plate is about 50 to 200, it is critical to use a trajectory optimization technique to reduce the total traveling time between colonies. Second, the scattering pattern is generated via the relative position of two entities: the laser and the colony. Therefore, one achieves a circularly symmetric scattering pattern when the center of the colony is close to the center of the incoming laser. This 2-D centering process can be done by calculating the geometric centroid of the scattering pattern relative to the imaging frame.
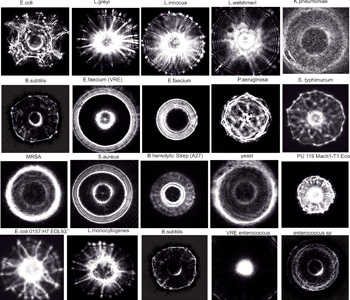 Forward-scatter patterns from bacterial colonies collected using ELS technology. Here we show optical signatures of colonies of different bacterial strains collected using ELS. Profiles are highly reproducible and require only one to two seconds per colony to obtain. There are a variety of profiles, depending on the size, shape, height and composition of the colony. Agar density, color and water content can also contribute to variations.
Forward-scatter patterns from bacterial colonies collected using ELS technology. Here we show optical signatures of colonies of different bacterial strains collected using ELS. Profiles are highly reproducible and require only one to two seconds per colony to obtain. There are a variety of profiles, depending on the size, shape, height and composition of the colony. Agar density, color and water content can also contribute to variations.
ELS’s strengths and limitations
ELS should have a significant place in the arsenal of bacterial identification techniques: It can identify organisms without additional biochemical tests or genetic tools, thereby reducing costs. Moreover, ELS is faster than all other current identification techniques—in some cases working in mere seconds.
For example, E. coli cultures typically take about one day to grow. Conventional biochemical analyses and PCR can take four to seven days to confirm identification. ELS, on the other hand, can be used immediately after the bacterial culture is grown. The scattering patterns are automatically compared to a library of known patterns, allowing for near-instant identification. If, by chance, the scattering pattern does not have a match in the library, we can at least conclude that something of interest is growing on the E. coli-specific nutrient medium and investigate further.
Although some critics of ELS have suggested that it is limited because it requires a colony to work—and that the organism of interest be culturable—the same is true of virtually all microbial detection and identification techniques available today.
ELS technology has been tested on tens of thousands of bacterial colonies and on hundreds of strains and subspecies. The measurements are highly reproducible and very robust. The images on the facing page are sequential ELS patterns from a single rectangular plate that were collected using our fully automated collection system. Furthermore, identifying patterns for many bacteria have been stored in our database, allowing us to rapidly classify many different species. As an example, the image to the right shows 24 actual scatter patterns. The fingerprint of an organism is a deconvolution of many features extracted from these unique and magnificent scatter patterns.
Cost is another potential advantage of ELS. A mass-produced ELS system could consist of inexpensive, off-the-shelf hardware, such as red lasers and low-resolution digital cameras available at consumer electronics stores. The system also indirectly saves money by requiring no reagents and a small amount of bench space in the typical laboratory.
No technology is perfect. ELS is limited in that it currently operates only with bacterial colonies, and—in order to be measured—these colonies need to be more or less transparent and approximately the size of the laser-beam diameter. We are experimenting with smaller beam profiles, which would allow us to evaluate smaller colonies earlier in their development. The first profiles we studied were 24-hour colonies. More recently, we have been able to assess many organisms at 12 hours using a fully robotic system, which can make measurements at any time. We anticipate that this could be reduced even further with more advanced optics and imaging tools. Naturally, only organisms that are culturable can be measured with ELS. Culture time can contribute to the ELS pattern and must be considered in developing the required classification database.
Applications of ELS in the food industry
According to the U.S. Centers for Disease Control and Prevention (CDC), it can take up to two weeks to identify a pathogen after a person initially reports having become sick after eating contaminated food. Conventional physiological and serological methods require two to seven days to identify the pathogen because of the multiple steps involved in sample preparation and the need (in most cases) to establish a pure culture to work from. During this time, pathogens can spread among the public. Immunologically compromised individuals are at particular risk because they face greater health consequences from an infection.
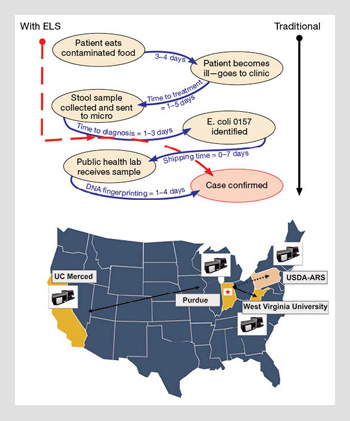 Bacteria identification with ELS and traditional methods. The U.S. Centers for Disease Control and Prevention (CDC) estimates that pathogen identification can take up to two weeks with traditional methods. ELS techniques can shorten this wait period. Laboratories can connect and compare results with a networked database. (Top) Modification from the CDC website showing the current traditional pathway for identifying pathogens from food. The dotted line on the left shows the shortcut that ELS technology offers by identifying the colony immediately after the ELS pattern is transmitted. (Bottom) The current distribution of networked ELS instruments in the U.S.A. With this system, early warning signals can be initiated using electronic fingerprints rather than waiting for physical distribution of a culture.
Bacteria identification with ELS and traditional methods. The U.S. Centers for Disease Control and Prevention (CDC) estimates that pathogen identification can take up to two weeks with traditional methods. ELS techniques can shorten this wait period. Laboratories can connect and compare results with a networked database. (Top) Modification from the CDC website showing the current traditional pathway for identifying pathogens from food. The dotted line on the left shows the shortcut that ELS technology offers by identifying the colony immediately after the ELS pattern is transmitted. (Bottom) The current distribution of networked ELS instruments in the U.S.A. With this system, early warning signals can be initiated using electronic fingerprints rather than waiting for physical distribution of a culture.
Therefore, within the process of monitoring and establishing countermeasures to the outbreak, timely intervention is critical. Even though final confirmation of such cases is usually decided at the national or international level before the public is informed, ELS could serve as a pre-screening tool to help contain the spread of disease.
In this scenario, if a local monitoring office were to have access to a national or international biodefense network ELS pattern database, and if each outbreak location were to have a portable scatterometer, it would be relatively easy to detect a possible outbreak and raise an alarm. By providing an early warning, ELS technology can improve the identification process and thus expedite patient treatment and slow transmission of foodborne pathogens.
Finally, the linking of databases across food safety inspection services would allow communities worldwide to identify common and unfamiliar pathogens much faster than can be achieved using present technology.
Opportunities for biodefense
ELS also shows great potential in the arena of biodefense—efforts to prevent or contain biological terrorism. Bioterrorism refers to the intentional release of harmful biological agents, such as bacteria, viruses or other toxins in either their naturally occurring or genetically engineered forms. Over the past decade, global governments and public health officials have put well-practiced protocols into place to address acts of bioterrorism. The goal is to rapidly detect mists or clouds of harmful molecules—which may be present in infinitesimal concentrations—and then launch a formal public health response. However, this process will work only in areas where detection systems are well distributed—which is not the case for most places in the world.
One of the least rehearsed bioterrorism scenarios, and the one for which the fewest protocols exist, would be an instance in which a group of terrorists simultaneously distributes modified pathogens with a severe and debilitating impact in several large cities around the world. No one would suspect the attack until large numbers of people begin to fall ill. Within hours or days, emergency wards would see more and more affected individuals.
Even in the best-case scenario, it would take up to 36 hours for body fluids to be sampled, sent to labs and reviewed. And that is assuming that the organism has not been genetically altered, in which case current detection systems may fail to identify it, significantly delaying public health and security responses. Furthermore, since no existing technology uses an electronically distributed fingerprint from a single colony, operational systems could not rapidly survey a global database for similar signatures as ELS can.
Clinical practice
Imagine this scenario: A person enters a hospital emergency ward with what appears to be food poisoning. The doctor collects a specimen from the sick person and sends it to the microbiology laboratory for analysis. Bacterial samples are plated on the first day and are not evaluated until the next day.
On the second day, researchers in the lab will decide whether further selective nutrient media and genotyping of the colonies are needed. It may take a third day before pure colonies can be identified and sensitivity tests conducted. However, identification may still not be possible, particularly if the organism is not a well-known pathogen with primers already included in the laboratory’s PCR protocols or fingerprinting tests. In the meantime, the sick person is most likely receiving a general treatment that may or may not be effective in alleviating symptoms or inhibiting the pathogen.
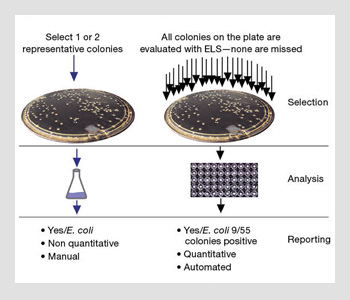 Traditional methods vs. ELS technology for identifying bacteria. If ELS is used, all colonies derived from the initial specimen culture are processed, reducing the possibility of missing organisms.
Traditional methods vs. ELS technology for identifying bacteria. If ELS is used, all colonies derived from the initial specimen culture are processed, reducing the possibility of missing organisms.
In a more troubling scenario, the pathogen might be missed entirely. As shown in the image to the right, a laboratory technician selects a single colony as being representative of a particular type of pathogen of interest (such as a colored colony on selective agar), and he or she then sub-cultures it and subjects it to sensitivity tests. What happens if the technician chooses the wrong representative colony? ELS techniques eliminate this potential for human error. Every colony is automatically selected, analyzed and classified without human intervention.
ELS technology has the unique potential to rapidly identify and distinguish between the electronic signatures or fingerprints of high-priority agents like anthrax, botulism and plague versus those that cause more common threats such as water and food contamination, ricin toxin and viral encephalitis. It allows us to quickly assess the potential risk and initiate the appropriate response even before the precise identity of that organism is known. In other words, if an unknown ELS pattern arises, it can be defined without any other information through a comparison to all defined ELS patterns in the networked database. This works because ELS produces unique mathematical patterns for bacterial colonies.
In summary
ELS is a sufficiently advanced technology to thwart sophisticated acts of bioterrorism. Its ability to monitor the signature patterns of common foodborne pathogens across the community brings an added dimension to food pathogen monitoring. A large networked database of bacterial signatures based on biophysical rather than genetic properties, paired with a technology utilizing machine-learning tools for emerging pathogen detection, creates a unique opportunity to improve monitoring in food and water production, clinical settings and bioterrorist events.
The technology used to feed information into this database would be based on ELS patterns, from which unique identifying patterns for most known bacteria would be created and distributed around the world. We anticipate that, within several years, this method may become one of the most important technologies we have in the microbiological world. Who would have thought that these beautiful optical patterns would originate from bacterial colonies that have been evaluated the same way for over 100 years? Louis Pasteur would be impressed!
This work was funded by NIH grant 1R56AI089511from NIAID and the USDA via the Center for Food Safety Engineering at Purdue University.
J. Paul Robinson is director of the Purdue University Cytometry Laboratories in West Lafayette, Ind., U.S.A. B.P. Rajwa, E. Bae, V. Patsekin, A.M. Roumani, A.K. Bhunia, J.E. Dietz and V.J. Davisson are with Purdue University. M.M. Dundar is with Indiana University–Purdue University in Indianapolis, Ind. J. Thomas is with West Virginia University in Morgantown, W.Va. E. Daniel Hirleman is with the University of California, Merced, Calif.
References and Resourcess
>> R. Atkinson et al. Morbidity and Mortality Weekly Report. 55, 1042 (2005).
>> B. Bayraktar et al. J. Biomed. Opt. 11, 34006 (2006).
>> CDC Timeline for Reporting of E. coli Cases. (2006).
>> P. Banada et al. Biosens. Bioelectron. 22:1664-71 (2007).
>> E. Bae et al. Appl. Opt. 46, 3639 (2007).
>> E. Bae et al. J. Biomed. Opt. 13, 014010 (2008).
>> P.P. Banada et al. Biosens. Bioelectron. 24, 1685 (2009).
>> E. Bae et al. J. Biomed. Opt. 15, 045001 (2010).
>> B. Rajwa et al. Cytometry A. 77, 1103 (2010).
>> Baby BugBuster Project: A low-cost diagnostic instrument for creating bacterial identifications.
>> A fully automated bacterial scatter technology using robotics.