 [Getty Images]
[Getty Images]
In early 2011, two scientists walked the cobblestone streets of Old San Juan, the historic district of Puerto Rico’s capital. Recently introduced by a mutual colleague at a nearby microbiology conference, the pair soon fell into a conversation about a mysterious protein called Csn1. The protein helped bacteria to defend against viruses, but it wasn’t clear how it worked. Some speculated that it acted like genetic scissors, cutting up the invading viral DNA and neutralizing the threat. But no one had demonstrated that yet.
 Biochemist Jennifer Doudna and microbiologist Emmanuelle Charpentier, pictured in 2016. The pair was awarded the 2020 Nobel Prize in Chemistry for their work developing the CRISPR–Cas9 gene-editing system. [picture alliance via Getty Images]
Biochemist Jennifer Doudna and microbiologist Emmanuelle Charpentier, pictured in 2016. The pair was awarded the 2020 Nobel Prize in Chemistry for their work developing the CRISPR–Cas9 gene-editing system. [picture alliance via Getty Images]
The pair of scientists—Jennifer Doudna, based in California, and Emmanuelle Charpentier, working in Sweden—set out to uncover the inner workings of Csn1 and its role in the larger bacterial immune system. They found that the protein did indeed cleave foreign genetic material and established that it could form part of an incredibly powerful, versatile technique to cut DNA at any location desired within the genome.
The long-distance collaboration of Doudna and Charpentier on Csn1—now known as Cas9—led to a Nobel Prize-winning discovery whose impact rivals history’s biggest biomedical breakthroughs. Alongside tools like DNA sequencing and polymerase chain reaction, humanity now has at its fingertips CRISPR–Cas9, a genome-editing technique that allows researchers to alter the DNA of organisms at will.
While the CRISPR–Cas9 system has already achieved revolutionary results, how to control its action in biological and eventually clinical settings remains a big open question. And increasingly, researchers favor using light as the control switch—to turn genome editing on and off, to reduce the probability of off-target effects and to put CRISPR–Cas9 to work in exactly the right spot.
While CRISPR–Cas9 editing has achieved revolutionary results, how to control its action remains an open question. Researchers increasingly favor using light as the control switch.
“In general, we deliver CRISPR–Cas9 to repair or edit certain genes without any controls. Once you deliver this treatment systematically, it will distribute everywhere in the body,” says Yuan Ping, a professor of pharmaceutical sciences at Zhejiang University, China. “But if you shine light directly on the liver, genome editing only happens there. In the era of precision medicine, I think that light can provide this opportunity to impart ... precision to health care in the future.”
Genome-editing powerhouse
As the name implies, the CRISPR–Cas9 system has two components, originally discovered as part of a simple immune system in E. coli bacteria. CRISPR—an acronym for “clustered regularly interspaced short palindromic repeats”—constitutes small fragments of viral genetic material that the bacterium embeds in its own DNA to recognize viral intruders. Cas9 is an enzyme used in the system to cut up and disable the DNA in viruses so identified. (The “Cas” in Cas9 stands for “CRISPR-associated protein.”)
In the bacterial system, the CRISPR section of the genome, which contains material embedded in the bacterial DNA after previous viral encounters, is transcribed into a long, single-stranded RNA molecule. Short RNAs called trans-activating CRISPR RNAs (tracrRNA) are then created that fit into the repeated sequences like puzzle pieces, and that are tied to the Cas9 enzyme.
The longer RNA originally transcribed from the CRISPR DNA section is then cut by a different enzyme into sections—so-called CRISPR RNAs (crRNAs)—that contain the genetic information needed to identify each unique virus encoded in the CRISPR section. When one of the individual crRNAs fits into a region of viral DNA, signaling an attack by an incoming virus, the tracrRNA and Cas9 work together to identify and chop up the viral DNA, stopping the invading genetic material in its tracks.
Doudna and Charpentier simplified the bacteria’s natural system by combining crRNA and tracrRNA into a single molecule, which they named guide RNA. This allowed scientists to create a guide RNA that matches any section of DNA where a double-stranded break should be made—in principle, in any organism. Cutting at any desired location within the genome also means that removing old genes and inserting new ones can be performed with ease. (See “How CRISPR–Cas9 editing works,” below.)
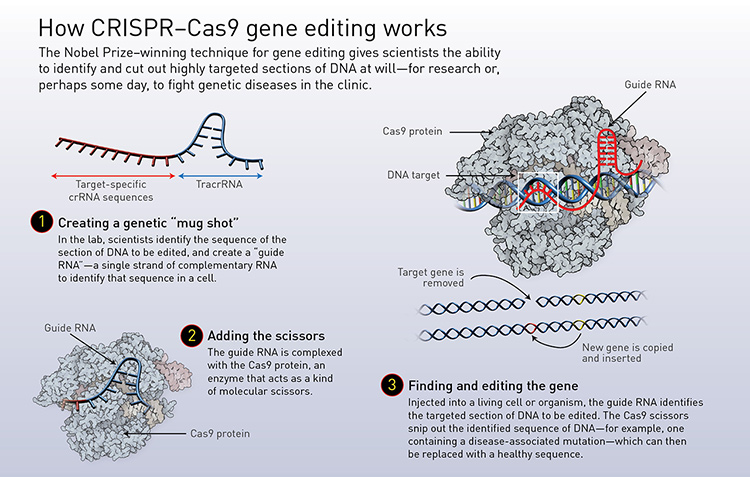 [Illustration by Phil Saunders] [Enlarge graphic]
[Illustration by Phil Saunders] [Enlarge graphic]
In the eight years between their fateful meeting in Puerto Rico and their receipt of the 2020 Nobel Prize in Chemistry, the groundbreaking work of Doudna and Charpentier, as well as others, has led to a tsunami of research on CRISPR-based systems and how they might be used to easily modify genomes in plants, animals and even humans. Targeted genome editing with CRISPR-Cas9 has already created more pathogen-resistant and productive crops, changed the DNA of cells and animals in the laboratory for basic research, expedited livestock breeding and engineered new antimicrobials. Clinical trials for the treatment of some genetic disorders in humans are in early stages.
“The first reports with respect to the clinics are extremely promising,” says Dominik Niopek, an assistant professor at the Technical University of Darmstadt, Germany. “It appears that the system works very robustly in all different kinds of settings, and I think that’s the most notable property of it. It’s really almost trivial to adapt it to different organisms, have it be functional in different kinds of organs, and so forth.”
In search of an “on/off switch”
Despite their power and flexibility, one thing that CRISPR-based systems currently lack is a built-in on/off switch. After Cas9 has done its job, it retains the ability to snip DNA, and unintended, prolonged Cas9 activity has been associated with DNA cleavage in the wrong places—so-called off-target effects. Such effects “are often a concern by scientists when this technology is used for biomedical applications, especially for treating diseases,” according to Ping, who compares off-target activity to an overdose of a medication.
Elevated Cas9 activity has also been associated with chromosomal translocations, in which part of one chromosome attaches to another, and general damage to DNA, or genotoxicity. Because off-target effects often occur at a slower rate than on-target editing, researchers began to search for ways to shut off CRISPR–Cas9 after its job was done, which would also prevent a patient’s immune system from attacking Cas9 and avoid further alteration of the genome in cases that involved the editing of eggs or sperm.
“If I can control gene function and protein function with light, I can convey similarly high levels of spatiotemporal control to treatments,” says chemist Alexander Deiters.
To put a leash on Cas9, some researchers have experimented with chemical methods that employ tailored small molecules to activate or inhibit Cas9 activity. But these lack spatial specificity and could disrupt crucial processes in the edited cells. Optical control knobs, on the other hand, boast spatial and temporal precision, noninvasiveness and, in some cases, reversibility.
“If I can control gene function and protein function with light, I can convey similarly high levels of spatiotemporal control to treatments,” says Alexander Deiters, a professor of chemistry at the University of Pittsburgh, USA. “That is interesting because then you can make a strong argument that there may be minimized off-target effects.”
Light-based controls broadly fall into two main categories. Optogenetic control generally involves genetically modifying the CRISPR–Cas9 system or its components to include proteins that are controllable with light. Optochemical control uses light to regulate special molecules—often in the form of a photoswitchable “molecular cage” around the molecule—that can activate or inhibit Cas9 or guide RNA functions. (See “Optochemical versus optogenetic control,” below.)
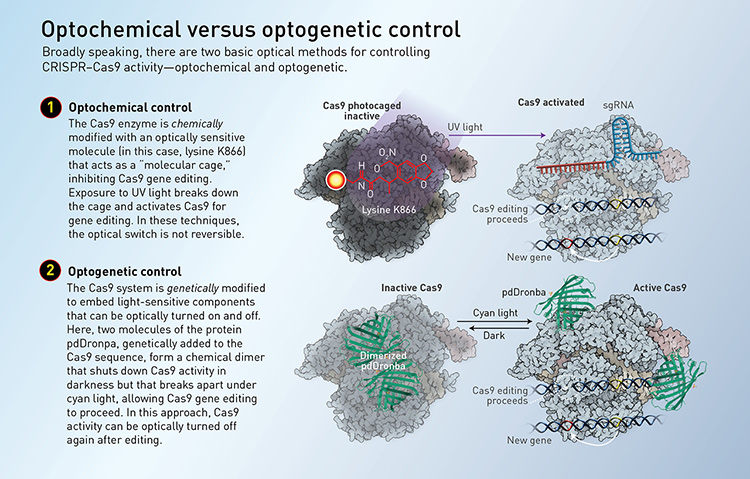 [Illustration by Phil Saunders] [Enlarge graphic]
[Illustration by Phil Saunders] [Enlarge graphic]
In 2015, two research groups independently reported optogenetic control of CRISPR–Cas9 for the first time, building a photoactivatable transcription system that uses the system, plus blue light, to target and turn on expression of specific genes. The goal of the work was not to control genome editing, but to ease laboratory studies in areas such as developmental biology. Since those initial studies, however, many other groups have come forward with both optogenetic and optochemical methods providing multiple avenues for controlling genome editing with light.
Targeting Cas9 activity
On the optochemical side, for example, Deiters and his colleagues, later in 2015, engineered a light-activated Cas9 for controlling genome editing. The team identified an amino acid within Cas9 that plays an essential role in its activity, and applied a molecular cage to inhibit its function. The cage is broken with exposure to ultraviolet light (365 nm); once that happens, Cas9 is free to go about its usual business.
“We did a sort of unnatural amino acid scan, where we inserted a photocaged lysine instead of a natural lysine in all these different sites, and we found one site where the photocaging group shut down the function of Cas9. Then we could restore it through a brief irradiation with UV light,” Deiters says. “We demonstrated that we could use this to achieve spatial and temporal control of Cas9 in mammalian cells.”
 Illustration of CRISPR-Cas9 gene editing complex. [J. Gaertner/Science Photo Library]
Illustration of CRISPR-Cas9 gene editing complex. [J. Gaertner/Science Photo Library]
A team led by Moritoshi Sato at the University of Tokyo, Japan, meanwhile, discovered a way to exert optogenetic control of Cas9 activity around the same time as Deiters’ optochemical study. The Japanese researchers engineered a split Cas9 consisting of two fragments, with special domains that join together in response to blue-light irradiation (450 nm). The domains consisted of a plant-based cryptochrome (a photoreceptor that serves to initiate flowering in the presence of blue light) and its binding partner.
Upon exposure to light from a blue LED source, the researchers found that genome-editing activity switched on. Extinguishing the light stopped all activity, as the Cas9 broke into pieces once more. The team also illuminated cells with slit-patterned blue light using a photomask, with the width of the slits set at 2 mm. The cells were programmed so that a double-stranded break in a certain targeted DNA sequence would cause the expression of green fluorescent protein. After 24 hours, the cells glowed green in the same pattern as the photomask, indicating that the team had achieved spatial control of genome editing with light.
More recent approaches have aimed to improve the simplicity, efficiency and clinical applicability of optical control methods. A team led by Michael Lin, an associate professor of neurobiology at Stanford University, USA, created a single-component optogenetic system in 2017 that could easily be applied to different Cas9 species. A molecular attachment—the photodissociable green fluorescent protein pdDronpa—converts Cas9 from a circular caged conformation in the dark to a linear open conformation in the presence of 500-nm (cyan) light.
“Instead of having two parts, it’s just one protein, and we make that protein able to bind to its DNA target in a light-dependent manner. We did that by fusing two domains to the CRISPR–Cas protein that bind to each other in the dark, but they’re dissociated by light,” says Lin. “So in this way, combining in the dark by these two domains actually blocks the DNA binding activity of the CRISPR Cas protein, whereas the dissociation of these domains by light allows the protein to bind to DNA.”
This approach produced three new photoswitchable Cas9 variants, two of which were employed in an experiment to test light-induced editing and transcription of different genes within the same cells. In the experiment, one variant targeted editing of a certain gene, while the other induced transcription and expression of a red fluorescent protein. After 48 hours of illumination, the cells demonstrated simultaneous optical induction of both gene editing and gene transcription.
Beyond Cas9: RNAs and anti-CRISPR proteins
A handful of research groups have looked at light-based controls for other parts of the CRISPR–Cas9 system. In 2020, Deiters and his colleagues applied their optochemical photocaging method to the system’s guide-RNA component. The team found that exposure to 365-nm UV light turned on genome editing with high spatiotemporal specificity in both mammalian cells and zebrafish embryos.
In 2019, the laboratory of Xinjing Tang, a professor of pharmaceutical sciences at Peking University, China, designed and constructed a photocaged crRNA by coupling it with a photolinker to a vitamin E molecule that significantly inhibited DNA cleavage. After a few minutes of 365-nm irradiation, the vitamin E molecule and photolinker completely detached from the crRNA, allowing normal CRISPR–Cas9 activity to proceed.
Taking an even less conventional path, Niopek and his colleagues turned their attention toward proteins found in bacteriophages that act as counter-defense agents against the bacteria’s immune system. His laboratory pioneered this unique optogenetic method back in 2018 and continues to refine it for greater versatility and ease of use.
“We had actually been thinking about natural systems that impair or control CRISPR–Cas activity, and it turns out that there is a very interesting class of proteins called anti-CRISPR,” says Niopek. “What we have been exploring is whether we can control these anti-CRISPR proteins with light.”
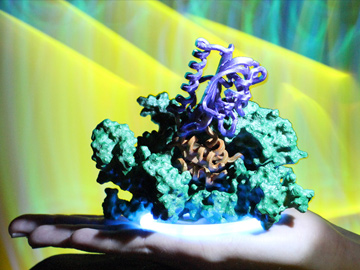 The lab of Dominik Niopek, Technical University of Darmstadt, Germany, is experimenting with a genetically engineered, light-activated “anti-CRISPR” protein (orange and purple domains in the 3D model above) that can photoswitchably neutralize Cas9 gene-editing activity. [Courtesy of D. Niopek]
The lab of Dominik Niopek, Technical University of Darmstadt, Germany, is experimenting with a genetically engineered, light-activated “anti-CRISPR” protein (orange and purple domains in the 3D model above) that can photoswitchably neutralize Cas9 gene-editing activity. [Courtesy of D. Niopek]
In nature, anti-CRISPR proteins avoid destruction of the bacteriophage’s DNA by interacting with crucial subunits of the Cas protein, preventing it from binding to or cleaving the invader’s DNA. Niopek and his colleagues leveraged this existing process for the conditional control of CRISPR–Cas9 by engineering artificial, photoswitchable anti-CRISPR proteins. They fused blue-light photoreceptor domains from the common oat to AcrIIA4, a bacteriophage-derived protein that blocks Cas9 DNA binding and activity.
Illumination of samples took place in a cell culture incubator affixed with six high-power, 460-nm blue LEDs. In the dark, AcrIIA4 behaved more or less normally in mammalian cells, significantly impairing Cas9 activity. When the LEDs were turned on, Cas9 activity recovered fully, suggesting that AcrIIA4 had been rendered nonfunctional.
“There is a simplicity in our approach in that people can use these light-dependent anti-CRISPR proteins we have developed and put them on top of what they already have established,” he notes. “We are currently working on light-dependent broadband anti-CRISPR proteins, which means you basically can use one single tool to control all different relevant Cas9 [variants].”
Eyeing clinical applications
Each method of optical control thus far demonstrated has its advantages and disadvantages. Any optochemical strategy involving photocages, for example, has the downside of being irreversible, since re-caging Cas9 or guide RNA is impossible. Some of the techniques require additional components, such as anti-CRISPR proteins or proteins that activate gene transcription, which adds complexity. The anti-CRISPR approach also requires the right ratio of anti-CRISPR proteins and Cas9. Too little anti-CRISPR will cause leakiness in the system and prevent full inhibition of Cas9. Too much will lead to over-inhibition, even after illumination.
Perhaps the biggest open question, however, is how to take these optical control techniques—most of which have been demonstrated only in cell cultures—and put them to work in animal models or the clinical realm. A major hurdle is the blue and UV wavelengths commonly used to activate CRISPR–Cas9, as such light can penetrate to only very superficial depths in biological tissues.
“Currently, optical control of CRISPR-based gene editing is only achieved mostly at the cellular level due to penetration limitation of UV or visible light,” says Tang. “More precise photomodulation and optical control of gene editing in animal models is still challenging.”
A few years ago, several groups started making strides toward addressing this issue by designing systems that are trigged by near-infrared light, in the 700-to-1000-nm range, which can penetrate to depths greater than 3 cm in biological tissues. One study, published in 2019 by researchers at three universities in China, employed lanthanide-doped upconversion nanoparticles (UCNPs) that could convert low-energy near-infrared radiation (980 nm) to high-energy UV light (365 nm).
The team modified the UCNPs with a silica layer where a photocleavable linker covalently anchors inactive Cas9. Upon exposure to light from a near-infrared laser, the UCNPs emit local UV light to cleave the linker and release the now-active Cas9 into cell nuclei for genome editing to take place. This optochemical technique was able to knock out a gene associated with cell proliferation in live, intact mice implanted with human lung cancer cells. Near-infrared illumination of the tumor led to slowed progression and decreased cell density when compared to a control group.
A second study from 2020, by a group led by Ping of Zhejiang University, showcased a system called nanoCRISPR that uses light from the second near-infrared optical window to heat up gold nanorods. Heat-inducible promoters take the cue and activate transcription of small DNA molecules that encode for Cas9.
“The second biological window usually refers to light with wavelengths greater than 1000 nm. But in biological systems, there are no proteins that can sense light in the near-infrared light region,” says Ping. “So we used nanomaterials as a mediator to transform light energy and either trigger Cas9 expression or inhibit its function.”
While some studies envision the prospect of therapeutic genome editing, most experts see optical control of CRISPR–Cas9 having the most impact as a research tool.
As with the previous study, tumors in mice were injected with nanoCRISPR, and then some animals were irradiated with a near-infrared laser (1064 nm) in order to knock out a gene that regulates cell duplication. Tumors that received irradiation shrank significantly after treatment, whereas tumors that did not receive irradiation grew rapidly.
Ping notes two disadvantages of the approach: it’s irreversible, and the safety of the gold nanorods in actual clinical use is unclear. “Designing degradable, optically responsive biomaterials,” he says, “is very important to move forward in this direction.”
The future of optical control
While these studies envision the prospect of therapeutic genome editing, most experts ultimately see optical control of CRISPR–Cas9 having the most impact as a research tool. Already, CRISPR-Cas9 and its variants have shown advantages over other genome-editing techniques for building genetically modified cell lines and organisms, the interrogation of dynamic gene function, and other biomedical research applications. Conditional control with light should only expand those horizons.
The possibility of providing safer gene therapies by incorporating optics is not out of the question, although simpler ways of limiting off-target effects are being tested that will likely reach the clinic first. For example, several engineered Cas9 variants have been developed that lead to enhanced specificity and discrimination of the targeted DNA.
Optical control does successfully shorten the duration of Cas9 activity, however, which should reduce off-target effects. And nanoCRISPR and other methods have demonstrated minimized non-specific editing in sites with the greatest potential for off-target activity. Even so, more studies are needed to determine whether optical control lowers off-target effects at the whole-genome level, and to overcome potential safety concerns and side effects.
Still, it seems clear that light-based control is here to stay as part of the CRISPR–Cas9 toolkit. “I am optimistic for the future of optical control of CRISPR,” says Tang. “At the least, it can be a powerful tool in biological research. In addition, if we could solve the problem of wavelength of light irradiation and improvement of CRISPR gene editing efficiency in general, it will be possible [for it] to be used in medical practice.”
Meeri Kim is a freelance science journalist based in Los Angeles, CA, USA.
For references and resources, go online: www.osa-opn.org/link/crispr-optics.
